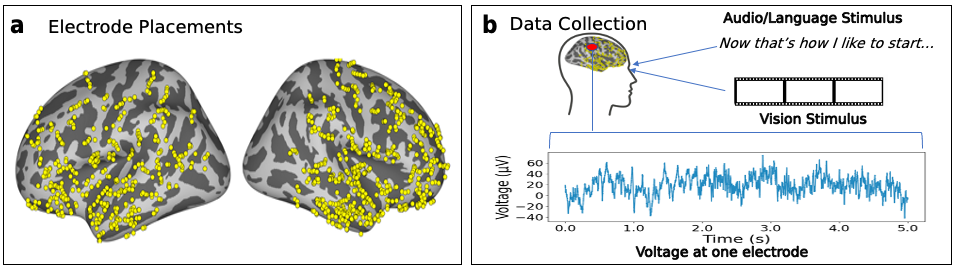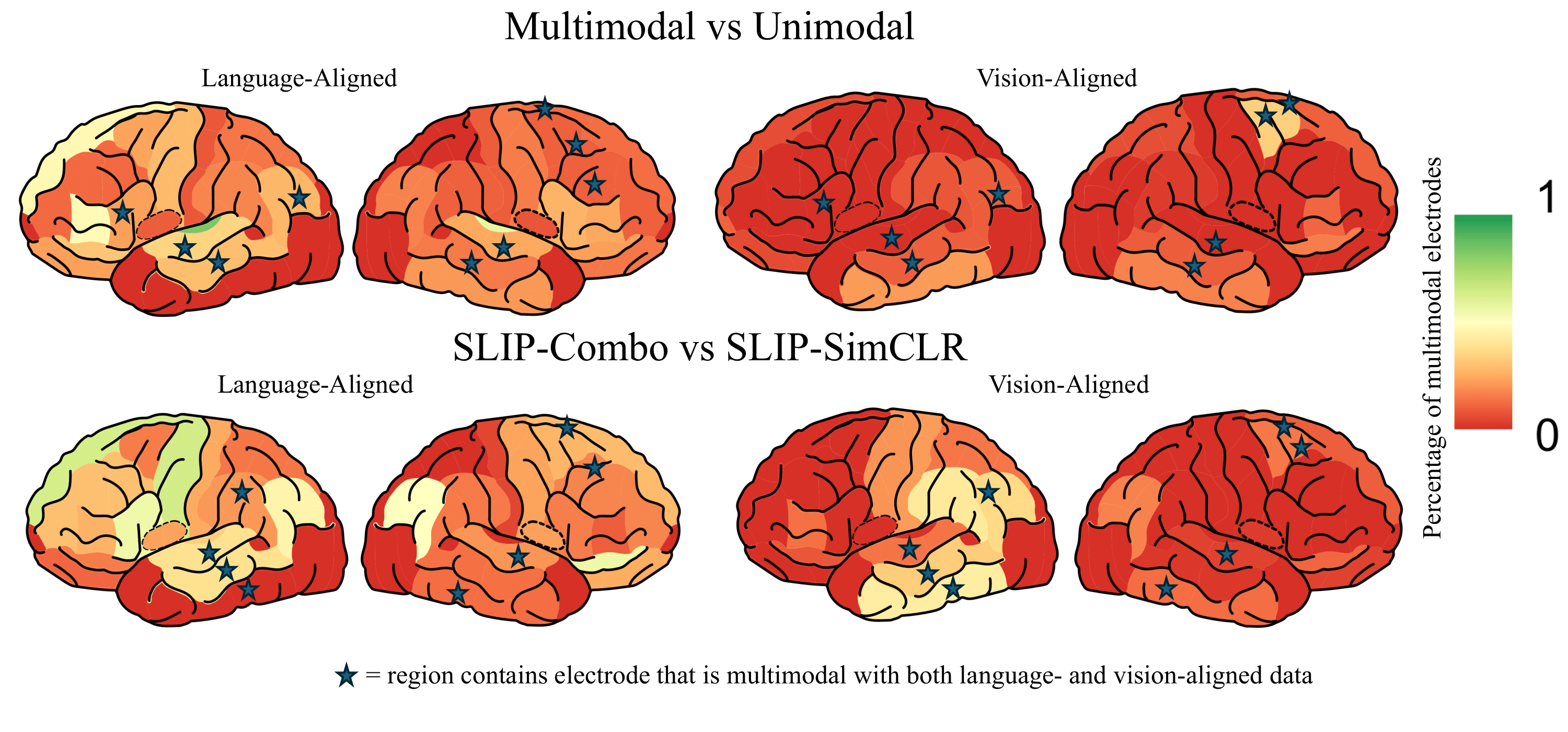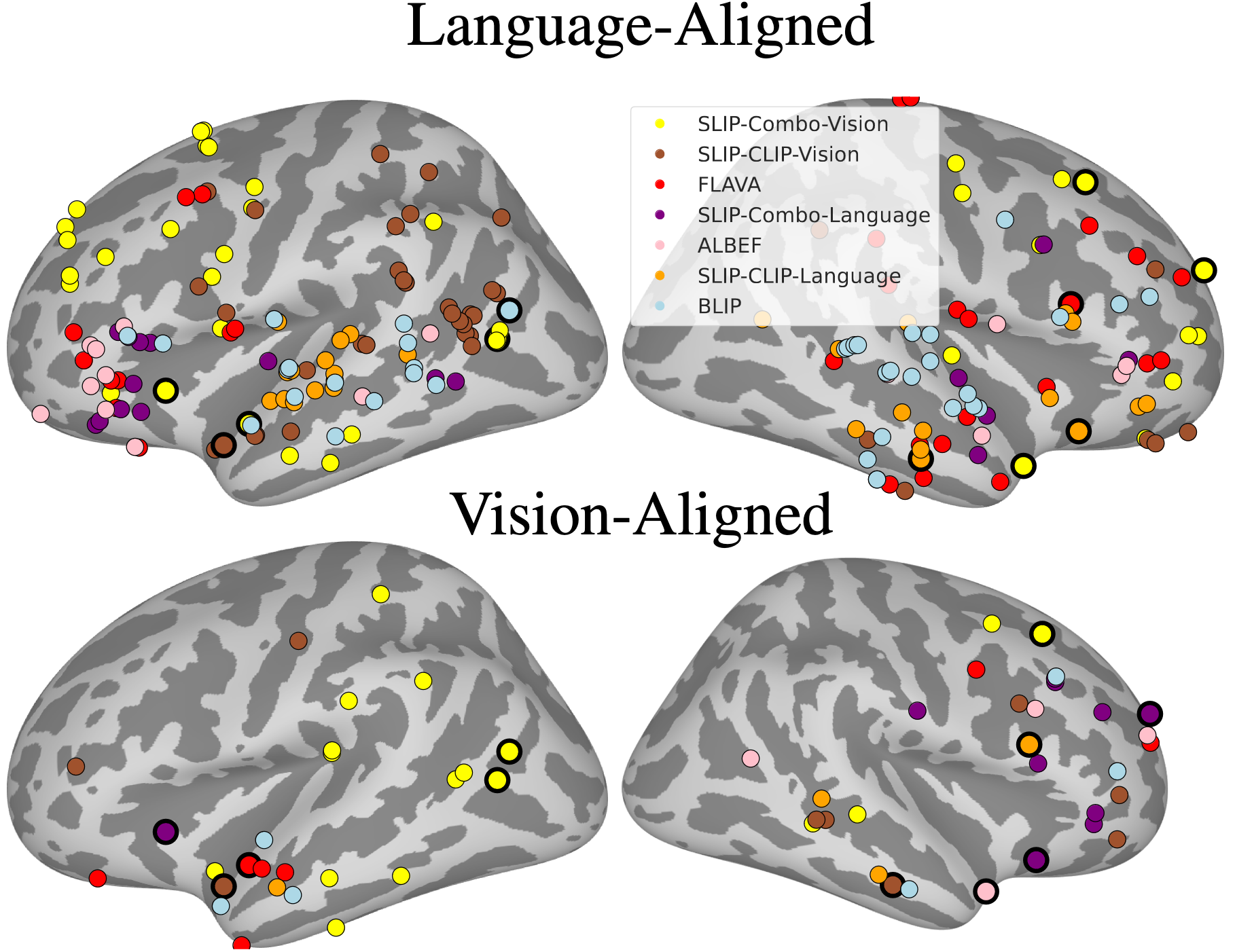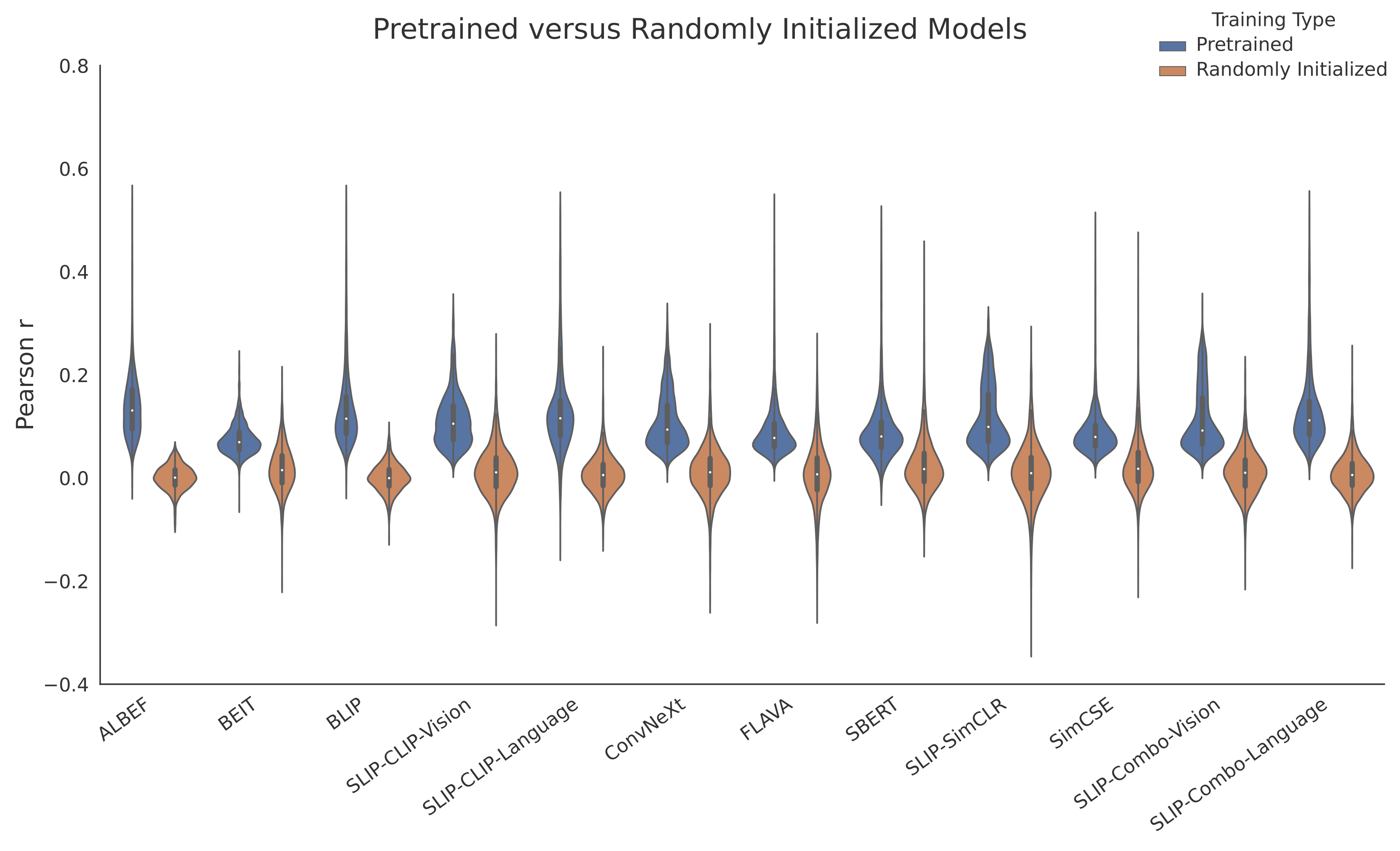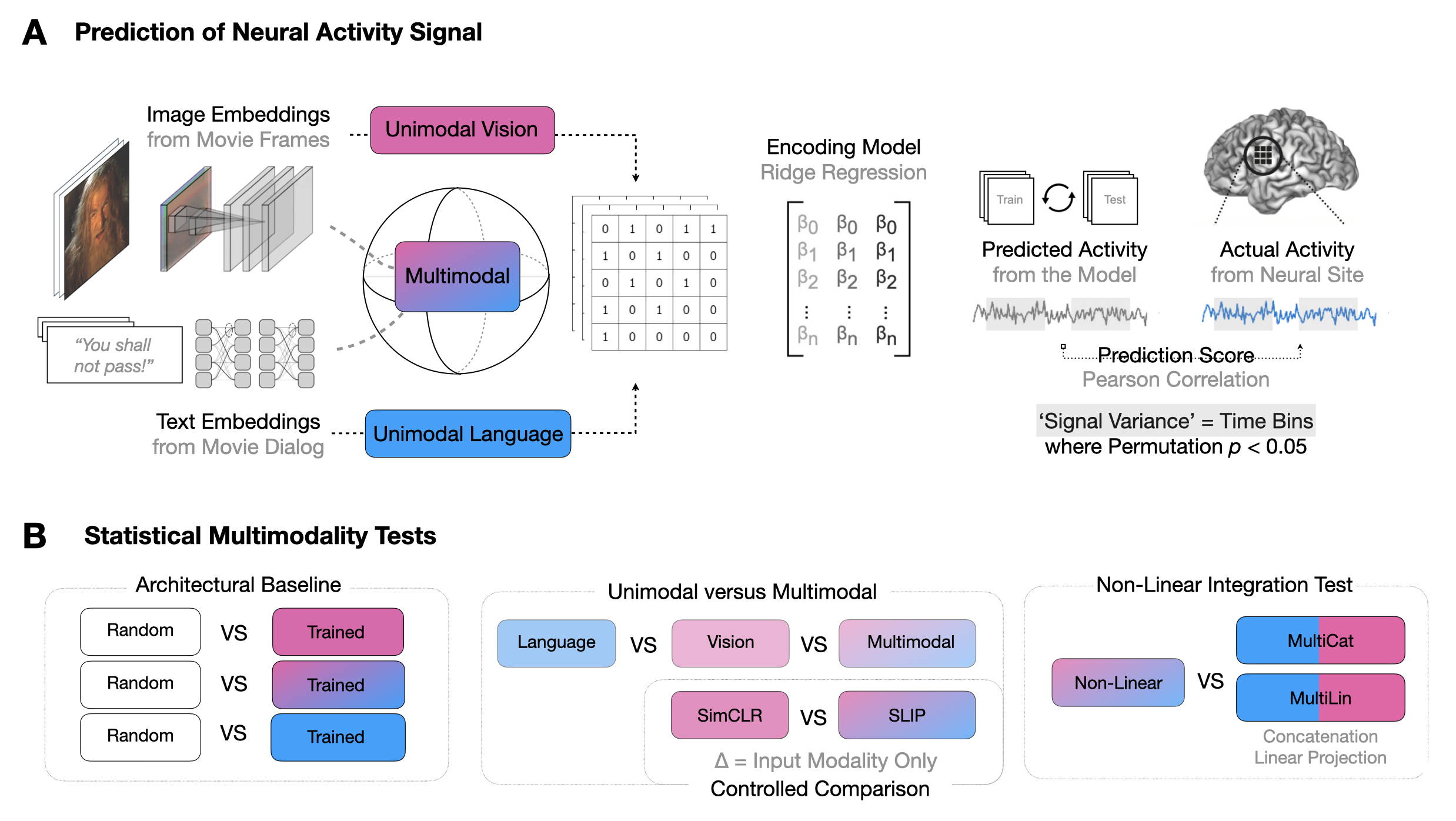
Multimodal Integration in the Brain Overview. (A) We parse the stimuli, movies, into image-text pairs (which we call \emph{event structures}) and process these with either a vision model, text model, or multimodal model. We extract feature vectors from these models and predict neural activity in 161 25ms time bins per electrode, obtaining a Pearson correlation coefficient per time bin per electrode per model. We exclude any time bins in which a bootstrapping test (computed over event structures) suggests an absence of meaningful signal in the neural activity target in that bin. We run these regressions using both trained and randomly initialized encoders and for two datasets, a vision-aligned dataset and language-aligned dataset, which differ in the methods to sample these pairs. (B) The first analysis of this data investigates if trained models outperform randomly initialized models. The second analysis investigates if multimodal models outperform unimodal models. The third analysis repeats the second holding constant the architecture and dataset to factor out these confounds. A final analysis investigates if multimodal models that meaningfully integrate vision and language features outperform models that simply concatenate them.